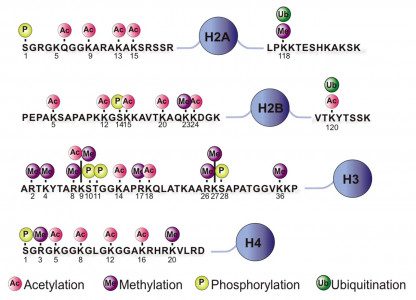
Epiproteomic Histone Modification Panel
The Epiproteomic Histone Modification Panel (EHMP) provides rapid, low-cost profiling of histone modification states within a sample. While not providing the depth and localization of ChIPSeq analysis, the EHMP allows researchers to generate hypotheses for future study. At present, these 92 modification states are assayed. (More will be added in the near future).
Sample requirements for histone assay:
1.) Cells Requirement: 1 – 5million
2.) Tissue Requirements: 25 – 100mg
| H1.4: K25UN | H1.4: K25AC | H1.4: K25ME1 | H1.4: K25ME2 | H1.4: K25ME3 | H2A: K5UN | H2A: K5AC | H2A: K9UN | H2A: K9AC |
| H2A: K36UN | H2A: K36AC | H2A1: K13UN | H2A1: K13AC | H2A1: K15UN | H2A1: K15AC* | H2A3: K13UN | H2A3: K13AC | H2A3: K15UN |
| H2A3: K15AC | H3R2UN: K4UN | H3R2UN: K4AC | H3R2UN: K4ME1 | H3R2UN: K4ME2 | H3R2UN: K4ME3 | H3R2UN: Q5UN | H3R2UN: Q5ME1 | H3: K9UN |
| H3: K9AC | H3: K9ME1 | H3: K9ME2 | H3: K9ME3 | H3: K14UN | H3: K14AC | H3: K18UN | H3: K18AC | H3: K18ME1 |
| H3: Q19UN | H3: Q19ME1 | H3: K23UN | H3: K23AC | H3: K23ME1 | H3: R42UN | H3: R42ME2* | H3: R49UN | H3: R49ME2* |
| H3: Q55UN | H3: Q55ME1* | H3: K56UN | H3: K56AC | H3: K64UN | H3: K64AC | H3: K79UN | H3: K79AC | H3: K79ME1 |
| H3: K79ME2 | H3: K79ME3 | H3: K122UN | H3: K122AC | H3.1: K27UN | H3.1: K27AC | H3.1: K27ME1 | H3.1: K27ME2 | H3.1: K27ME3 |
| H3.1: K36UN | H3.1: K36AC | H3.1: K36ME1 | H3.1: K36ME2 | H3.1: K36ME3 | H3.3: K27UN | H3.3: K27AC | H3.3: K27M* | H3.3: K27ME1 |
| H3.3: K27ME2 | H3.3: K27ME3 | H3.3: K36UN | H3.3: K36AC | H3.3: K36ME1 | H3.3: K36ME2 | H3.3: K36ME3 | H4: K5UN | H4: K5AC |
| H4: K8UN | H4: K8AC | H4: K12UN | H4: K12AC | H4: K16UN | H4: K16AC | H4: K20UN | H4: K20AC | H4: K20ME1 |
| H4: K20ME2 | H4: K20ME3 |
* Modification Available upon Request
AC=Acetylated, ME=Methylated, UN=Unmodified, and the number refers to the degree of modification. H3K9me3 indicated the trimethylation of lysine 9 of H3. Unmodified peptides are also included.
Related Publications
LaFave LM, Béguelin W, Koche R, Teater M, Spitzer B, Chramiec A, Papalexi E, Keller MD, Hricik T, Konstantinoff K, Micol JB, Durham B, Knutson SK, Campbell JE, Blum G, Shi X, Doud EH, Krivtsov AV, Chung YR, Khodos I, de Stanchina E, Ouerfelli O, Adusumilli PS, Thomas PM, Kelleher NL, Luo M, Keilhack H, Abdel-Wahab O, Melnick A, Armstrong SA, Levine RL “Loss of BAP1 function leads to EZH2-dependent transformation.” Nat Med, 2015, 21(11) p. 1344-9.
Summary | Full Text (PMC) | Full Text (DOI)
Zheng Y, Sweet SM, Popovic R, Martinez-Garcia E, Tipton JD, Thomas PM, Licht JD, Kelleher NL “Total kinetic analysis reveals how combinatorial methylation patterns are established on lysines 27 and 36 of histone H3.” Proc Natl Acad Sci U S A, 2012, 109(34) p. 13549-54.
Summary | Full Text (PMC) | Full Text (DOI)
Zheng Y, Thomas PM, Kelleher NL “Measurement of acetylation turnover at distinct lysines in human histones identifies long-lived acetylation sites.” Nat Commun, 2013, 4() p. 2203.
Summary | Full Text (PMC) | Full Text (DOI)
Pricing
https://proteomics.northwestern.edu/services/rates/
Instruments Used
Thermo Scientific TSQ Altis Triple Quadrupole Mass Spectrometer
Protocols
Download